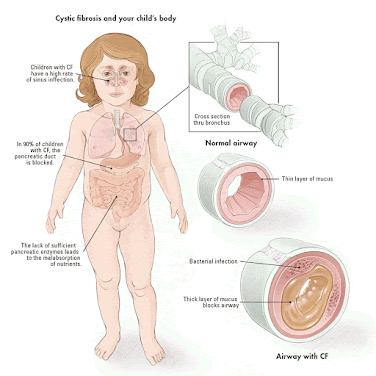
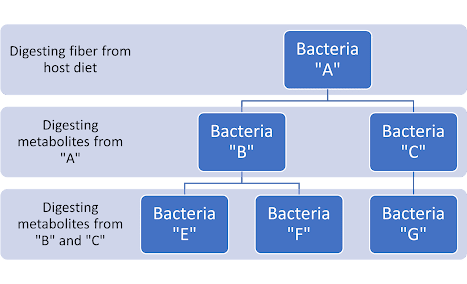
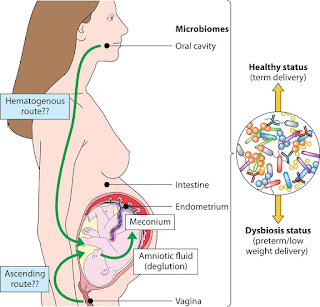
WEEK 7 Microbiome and Allergies
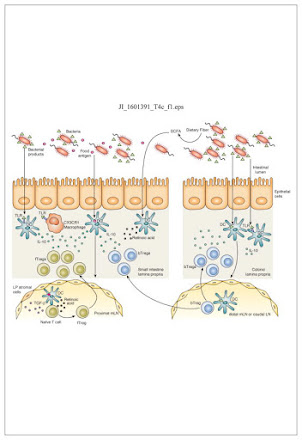
Presentation by ASU student Ainsley Chapman of review paper “The influence of the microbiome on allergic sensitization to food” Plunkett et al., J Immunol. 2017, 198(2): 581–589. doi: 10.4049/jimmunol.1601266. The Influence of the Microbiome on Allergic Sensitization to Food (wpmucdn.com) An increase in allergy prevalence in recent years was too great to be explained by genetics alone. The increase was most noticeable in developed countries concurrent with sanitation improvements, dietary changes, and antibiotic use. Dysbiosis caused by these changes may be to blame. Regulator T cells "Tregs", when triggered, calm and quiet the immune response. Byproducts of bacteria contain some of the triggers for Tregs. With changes in diet, antibiotic use and sanitation, the microbiome is altered. The host, now missing certain bacteria, is also missing the associated triggers for Tregs, leading to susceptibility to food and other allergies/sensitivities. Tregs are also found in oth